Scientists: Composition of the Vault
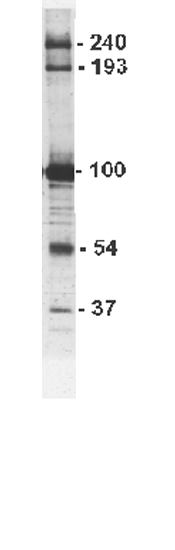
Since their discovery, vaults have been isolated from many different eukaryotic species including rat, mouse, rabbit, frog, electric ray, and monkey. SDS-PAGE analysis of vaults from higher eukaryotes reveals that the particle contains four major components. Three of these components are proteins and the fourth is an RNA. Thus the particle can be classified as a ribonucleoprotein particle. With a mass of 13-MDa and dimensions of ~72 x 41 nM, vaults are the largest cytoplasmic RNP known. Most of the structural work has been carried out on rat liver vaults . These vaults contain a protein of ~100 kDa (designated MVP for major vault protein) which makes up about 70% of the particle mass. Other proteins with apparent molecular masses of 240, 193 and 54 kDa are also seen in addition to the un-translated small RNA (vault RNA, vRNA, see below) which runs on SDS-PAGE at 37 kDa and is present in multiple copies. An SDS-PAGE gel of purified rat vaults is shown at the right, the vault component sizes in kDa are labeled.
Vaults from all species have an MVP of approximately 100 kDa which dominates the protein profile of the particle, however, only vaults from higher eukaryotes appear to have the high molecular weight (240 and 193 kDa) proteins while the ~54 kDa protein appears to be seen only in rat liver vault preparations. This protein was identified in 2002 as the La autoantigen. La is a single stranded RNA binding protein that may be involved in the delivery of vRNA to vaults (Kickhoefer et al., 2002). Vaults from the cellular slime mold Dictyostelium contain multiple related MVP isoforms of 94 kDa (MvpA) and 92 kDa (MvpB and MvpC) (Vasu et al., 1993, Vasu and Rome, 1995).
Vaults have not been detected in yeast, worm or flies (by database homology searches).
The vault RNA was first observed as a 37 kDa band on silver-stained SDS-PAGE gels that was not stained with Coomassie brilliant blue, suggesting that it might not be a polypeptide. This species was shown to be a small RNA; it could be extracted by phenol and was digested by ribonuclease but was resistant to proteinases and DNase. The RNA migrated as a single major species between the 5S and 5.8S ribosomal standards, on urea PAGE. The possibility remained that the 140-base vault RNA (vRNA) was a degraded remnant of an appreciably larger species and was generated by nucleases during the four-day vault purification process. Protein-specific anti-vault antibodies were used to immunoprecipitate vaults from 32P-labelled extracts of hepatoma cells. These immunoprecipitates, when phenol extracted and resolved on urea-PAGE, revealed a single labeled species with identical mobility as the RNA from purified vaults. This demonstrated that the RNA was not degraded during the long purification procedure by endogenous RNases, and also indicated that the vRNA was tightly associated with the vault protein (Kedersha and Rome, 1986a). Both results argue that the vRNA is an integral component of the vault particle. Attempts to sequence the vault RNA by Robert Searles were initially hampered by 3' heterogeneity. However, once Robert determined that the 3' end was frayed, direct RNA sequencing initially by Robert and later by Valerie Kickhoefer, revealed a sequence which was consistent with the base composition determined by HPLC. Val confirmed her direct RNA sequence by sequencing a genomic clone. She went on to demonstrate that the vRNA is transcribed by RNA polymerase III (Kickhoefer et al., 1993).