Scientists: Vault Assembly
The MVP is thought to be present in 96 copies per vault (see Structure). In many ways vaults are reminiscent of virus particles, in that they are very large assemblies that have a protein shell composed of multiple copies of a single protein and have a large central cavity. The expression of specific viral proteins in an insect cell expression system (baculovirus) has become an invaluable tool for virologists to investigate the assembly of virus particles. There are numerous examples where expression of a single viral protein was capable of directing the formation of virus-like particles, however, other studies required expression of multiple viral proteins.
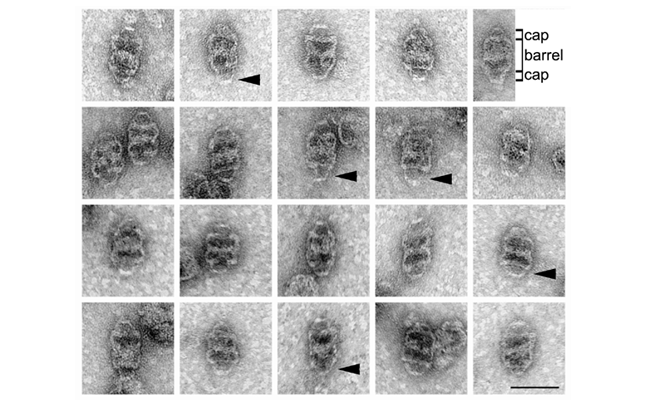
Currently nothing is known of the mechanisms by which vault particles are assembled within the cell. To gain insight into the mechanisms for vault assembly, the rat MVP was expressed in the Sf9 insect cell line using an insect cell viral vector. The results show that the expression of the rat MVP alone can direct the formation of particles that have biochemical characteristics similar to endogenous rat vaults and display the distinct vault-like morphology when negative stained and examined by electron microscopy (Stephen et al., 2001).
Although a considerable variation in particle morphology was observed, most recombinant vaults were similar in size and shape to purified rat vaults. These particles varied from 32-37 nm across to 59-65 nm in length and displayed the distinctive bifold symmetric vault central barrel with dual caps (for a gallery of these structures see the Figure below; see also a recombinant vault with the barrel and caps labeled, the arrowheads indicate the structural variation seen in the ends). These particles are the first example of a single protein polymerizing into a non-spherically, non-cylindrically symmetrical structure. Understanding vault assembly will enable the design of agents that disrupt vault formation and hence aid in elucidating vault function in vivo.