Scientists: Designer Vaults
In 2002 vault research proceeded in a new direction. The cryoEM reconstructions clearly demonstrated that the vault particle was a hollow capsule and the experiments of Stephen et al. (2001) indicated that this basic barrel-like structure could be formed solely by the assembly of multiple copies of MVP expressed in the baculovirus protein expression system. These findings led to the initiation of a new multidisciplinary collaboration at UCLA between groups in the David Geffen School of Medicine (Rome, Stewart, Eisenberg), the College of Letters and Science (Zink) and the School of Engineering and Applied Science (Dunn, Mobouquette). A proposal was funded by the National Science Foundation (NSF) for formation of a Nanoscale Interdisciplinary Research Team (NIRT).
The overall goal of this NIRT was to develop a flexible, targetable nano-capsule by exploiting the vault. Understanding how the vault capsule could be formed and manipulated in vivo and in vitro would allow molecular manipulations to encapsulate small molecules (drugs, sensors, enzymes, toxins etc.), lengthen and shorten capsule size, and target the engineered nanostructures to specific tissues, cells, or organelles via attachment to receptors, ligands, or fusogens.
Identification of a vault targeting "zip code"
Critical to engineering vaults was the development of a method to package foreign materials into the vault lumen. Accordingly, a strategy was developed to identify a vault targeting sequence, This grew from previous studies of the VPARP protein. This vault protein was identified using the yeast two-hybrid method employing MVP as bait (ref). The smallest MVP interacting VPARP clone encoded a 162 amino acid sequence found at the C-terminus of the full-length VPARP protein (Figure). We called this region of VPARP the INT domain (INT because it was responsible for interaction of VPARP with MVP). We also showed that a GST-INT fusion protein was sufficient to pull down MVP using a pull-down assay. To determine whether this domain could target a non-vault protein into vault particles, the cDNA encoding the firefly luciferase protein was fused onto the INT domain (Figure). Attachment of the INT domain to the C-terminus of the 60 KDa luciferase enzyme (LUC-INT) and expression in the baculovirus system resulted in a fusion protein that retained enzymatic activity. Co-expression of LUC-INT and MVP resulted in its assembly into the lumen of the vault. Cryo EM and single-particle image reconstruction revealed that the LUC-INT protein was packaged inside the vault resulting in additional density in two rings in the interior of the particle (Figure).
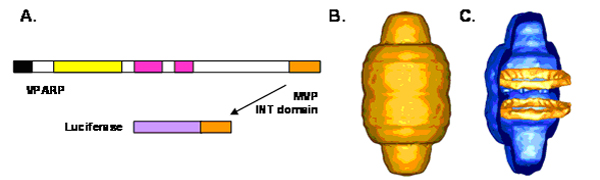
Figure.
(A) The C-terminal 162 amino acids of VPARP were fused to the C-terminus of luciferase. The luciferase-INT fusion protein (LUC-INT) retained enzymatic activity. (B) CryoEM of the MVP-LUC-INT vault reconstruction. (C) Difference map of the LUC-INT/MVP particles minus the MVP-only particles. The difference density is shown in orange superimposed over a half vault from the MVP-only reconstruction. The LUC-INT is seen as two rings of internal density shown in orange. From Kickhoefer et al. (2005).
In addition to targeting luciferase into the particle, we have also demonstrated that the INT domain can target the ~30 KDa green lantern protein (an analog of the green fluorescent protein, GL-INT) into the particle (Kickhoefer et al., 2005).