Big Kids: CryoEM Structure

A collaboration between the Rome lab and Phoebe Stewart's lab here at UCLA was initiated to carry out a cryoEM analysis of the vault. Lawrence Kong and Amara Siva led the reconstruction effort.
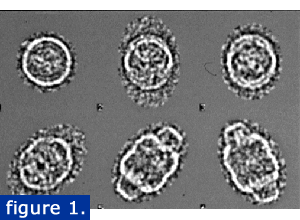
Raw cryo-EM vault images (figure 1).
Vaults were purified using a modification of the previously published method that included a CSCl gradient. Individual particles were quick frozen in liquid ethane and viewed by TEM. Images were captured with a digital camera and analyzed using IMAGIC software. The raw cryo-EM vault images are shown at right in various orientations from end to side views. Images that showed the particle broken or distorted were not included in the reconstruction.
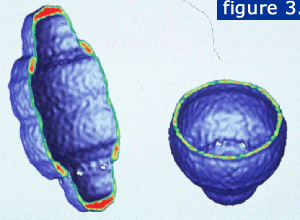
Over 1300 particle images were combined to calculate a 31 angstrom resolution structure of the vault. This structure (right) was generated with cyclic eightfold symmetry. It reveals a smooth outer shell with a barrel-shaped midsection and two protruding caps.
A slice through the particle reveals the hollow internal structure (below). The planes are displayed with the strongest density in red and the weakest density in green.
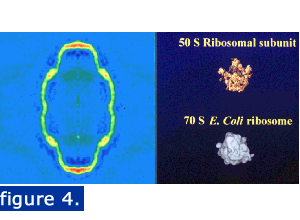
The vault is large enough to enclose particles such as ribosomal subunits. The image below shows a central density slice and two ribosomal subunits illustrated at the same magnification.