Big Kids: Composition of the Vault Particle
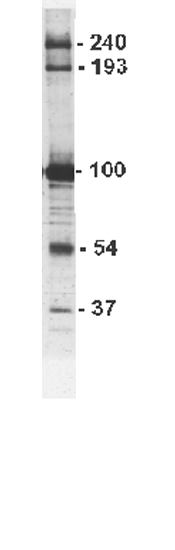
Nancy Kedersha worked out a method that permitted the purification of enough vaults to carry out a large number of biochemical studies. The purified material can be separated into it's component parts using a common biochemical method called SDS-PAGE which involves separation of the subunits of the particla on a gel. The gel can be stained to reveal the component parts of the particle. An example of such a stained gel is shown at the right.
Since their discovery, scientists in the Rome Lab at UCLA and elsewhere around the world (Kansas, Germany & Holland) have been able to isolate vaults from dozens of different higher organisms. Biochemical analysis of vaults from higher eukaryotes (rat, frog, sheep, etc..) reveals that the particle contains four major components. Three of these components are proteins (labelled 240, 193 and 100 in the figure to the right) and the fourth is an RNA (labelled 37). Because it is composed of protein and RNA, vaults can be classified as ribonucleoprotein (RNP) particles.
The total mass of the vault is 13 million Daltons and the dimensions of the particle are ~65 x 45 nM, making vaults by far the largest cytoplasmic RNP known.
The most abundant protein in vaults is a 100 kDa protein that comprises over 70% of the particle mass. This protein is refered to as MVP (for Major Vault Protein). Only vaults from higher organisms appear to have the high molecular weight (240 and 193 kDa) proteins while the 54 kDa protein appears to be seen only in rat liver and may be a stable breakdown product of the MVP.
The vault RNA (called vRNA) is an integral component of the vault particle. Direct RNA sequencing, initially by Robert Searles and later by Valerie Kickhoefer, was confirmed when Valerie isolated the gene for the RNA and sequenced it. She went on to demonstrate that the vRNA is transcribed (read from DNA) by an enzyme that produces other small RNAs called RNA polymerase III.